Recently, Functional Genomics of Plant Pathogenic Fungi Research Team from our college published a paper online in Science Advances with the title "Adaptive advantages of restorative RNA editing in fungi for resolving survival-reproduction trade-offs". This research is the latest progress on RNA editing function and adaptive advantages. Dr. Qi zhaomei is the first author of the paper, and Professor Liu Huiquan is the corresponding author.

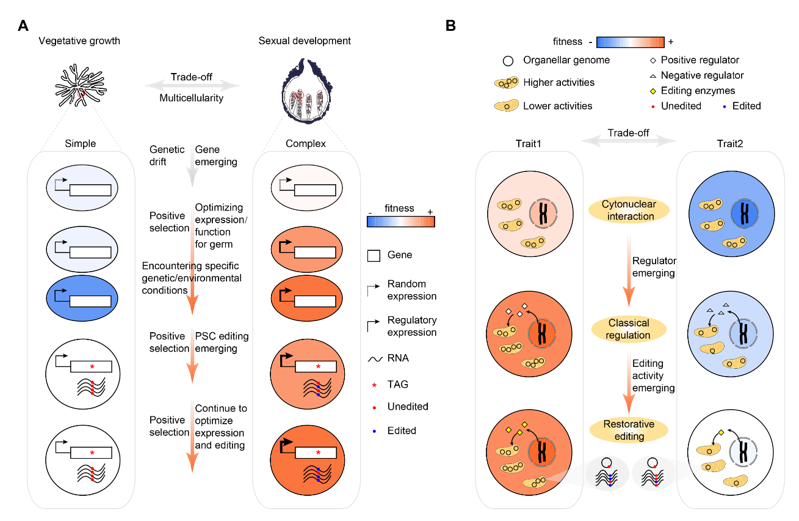
This study comprehensively used molecular biology, genetics, and evolutionary developmental biology methods to find that filamentous ascomycetes fungi have evolved to produce a large number of antagonistic pleiotropy genes, which are crucial for the reproductive stage of their life history, but are not beneficial to the stress adaptation of their non-reproductive stage hyphae growth. The A-to-I RNA editing mechanism in fungi drives these genes to undergo premature termination mutations, and repairs them at the RNA level during the reproductive stage, making them restore their function. This repair-type A-to-I RNA editing eliminates the survival-reproduction trade-offs caused by gene antagonism, giving fungi selective advantages. This study provides the first empirical evidence for the adaptive advantages of repair-type RNA editing, revealing the crucial role of RNA editing in the evolution and development of fungal complex multicellular structures, and enhancing our understanding of eukaryotic organism complexity. At the same time, this study shows that in the prevention and control of diseases caused by pathogenic fungi that prioritize reproduction, we cannot just focus on infection control, but should also take reproductive regulation into account.
Reproductive infructescences are the most complex multicellular structures produced by fungi, and the genetic principles underlying their complexity evolution remain unclear. This study found that filamentous ascomycete fungi have produced a large number of genes that originate de novo during infructescence evolution, and these genes play critical roles at different stages of infructescence development. This result highlights the importance of genetic innovation in the evolution and development of fungal complex multicellular structures, challenging the previously held view that gene (co-option) is the main force in complex multicellular evolution, while gene (de novo) evolution only plays a secondary role. The study also found that most of the new genes that play important functions in infructescence development are related to stress responses, and have antagonistic effects between survival and reproduction. These genes mediating survival-reproduction trade-offs have rarely been reported in fungi before, and their discovery is of great significance for understanding the formation of fungal complex multicellular structures and life history strategies.
Restorative RNA editing usually repairs gene mutations at the RNA level. Although these repair-type RNA edits are functionally important, there has been controversy about their adaptive advantages. Neutral evolution theory believes that the repair capacity of RNA editing allows harmful DNA mutations that should be eliminated by natural selection to accumulate. Correcting these "errors" is unnecessary complexity that does not provide any additional advantages. However, this study demonstrates that repair-type RNA editing in fungi can eliminate survival-reproduction trade-offs caused by gene antagonism, providing strong empirical evidence for the adaptive advantages of repair-type RNA editing. Because repair-type RNA editing occurs most frequently in eukaryotic cell organelles, this study provides an evolutionary model for the origin and maintenance of organellar RNA editing. In summary, this study provides new perspectives for understanding the origin and evolution of different RNA editing systems.
Research Scientists Jiang Cong and Wang Qinhuo as well as several graduate students from the team participated in this study. This research was supported by the National Key R&D Program "JIE BANG GUA SHUAI" project and the National Natural Science Foundation of China (NSFC) General Project.
Original Link: https://www.science.org/doi/10.1126/sciadv.adk6130